
Kent L. Hill, Ph.D.
4801A MSB, 148906
Affiliations
-Professor, Department of Microbiology, Immunology & Molecular Genetics
-Member: MBI, California NanoSystems Institute
-GBP Home Areas:
Immunity, Microbes & Molecular Pathogenesis;
Cell & Developmental Biology;
Biochemistry, Biophysics & Structural Biology
Research Interests
 |
 |
|
|
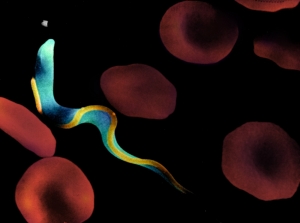
Figure 1. African trypanosome in the bloodstream. PMT = subpellicular microtubules, PFR = paraflagellar rod, FAZ = flagellum attachment zone. A) Scanning EM (provided by J.E. Donelson, University of Iowa)
|
Figure 2. Tsetse fly taking a bloodmeal. Figure provided by J.E. Donelson, University of Iowa |
| Research Program: Parasites in Motion, Mechanism and Biology of Flagellar Motility in Trypanosomes
My laboratory is investigating flagellar motility in African trypanosomes (Figure 1). These protozoan parasites cause a disease that is commonly called “African Sleeping Sickness”. They are transmitted to the bloodstream of their mammalian hosts through the bite of an insect vector, the tsetse fly (Figure 2). Once in the bloodstream, these highly motile, unicellular parasites burrow through the blood vessel endothelium and eventually invade the central nervous system, where they initiate a cascade of events that ultimately results in fatal sleeping sickness. The two general, long-term objectives of our research are: 1) Provide a better understanding of the cellular and molecular biology of trypanosomes and related kinetoplastid parasites, thereby facilitating the development of more effective treatments for the diseases caused by these organisms. These parasites are the source of mortality and morbidity in several million people worldwide and current treatment regimens are antiquated, costly and ineffective. 2) Exploit trypanosomes as a model system to investigate the function of the eukaryotic flagellum. As early diverging eukaryotes, trypanosomes have historically been a rich source for the discovery of novel biological phenomena that are subsequently found to occur in other eukaryotic organisms. Why Study Trypanosome Flagella? Cilia and flagella are evolutionarily-conserved organelles that protrude like small appendages from the surface of a cell. They are biological “nanomachines” that are present on most tissues of the human body and on many single-celled microbes. They perform motility, transport and sensory functions. Flagella are required for motility of human pathogens and defects in human cilia cause a variety of fatal and debilitating diseases. Infectious Diseases caused by pathogens that require cilia include: African sleeping sickness, Malaria, Epidemic Diarrhea and Trichomoniasis (the most common non-viral sexually transmitted disease in the world). These pathogens are responsible for mortality and morbidity in approximately 0.5 billion people world-wide. Heritable Human Diseases caused by cilia defects include: Hydrocephalus, Infertility, Left-Right Axis Defects, Eye Disorders, Polycystic Kidney Disease and Bardet-Biedle Syndrome (BBS). Therefore, in addition to addressing fundamental questions in cell biology, our research directly impacts efforts to understand and treat infectious diseases and genetic diseases in humans. Specific Research Projects include: I. Cell Motility. We have used the novel methodology of RNA-interference to generate inducible gene “knockdowns” in trypanosomes. One of the genes that we have “knocked-down” encodes a recently discovered protein that is essential for cell motility. Since trypanosomes are named for their hallmark auger-like motility (“trypanon” is Greek for auger), we have named this protein “trypanin”. Ongoing studies on this project include: i) Determining how cell motility influences parasite development and disease pathogenesis. ii) Identification and characterization of other components of the trypanosome’s motility apparatus. iii) Elucidation of the mechanisms by which trypanin controls cell motility. iv) Characterization of trypanin-related proteins that are present in other organisms. II. Protein Trafficking. Formation of the eukaryotic flagellum is dependent upon an evolutionarily-conserved protein targeting process termed Intraflagellar Transport (“IFT”), in which newly synthesized proteins are delivered from the cytoplasm into the flagellum. Recent work has led to the identification of IFT motors and other components of the IFT pathway. However, a unifying explanation for how flagellar proteins are targeted to the flagellum is unknown. We have identified a set of 30 novel flagellar proteins and are using these proteins to elucidate mechanisms of protein targeting to the flagellum. III. Additional areas of research include studies on a mammalian trypanin homologue and development of new methodologies for gene transfection and in situ gene tagging in trypanosomes. This latter project will benefit greatly from the recently completed trypanosome genome project. |
Selected Publications
1.Vélez-Ramírez, D. E., Shimogawa, M.M., Ray, S., Lopez, A., Raratpisheh, S., Langousis, G. Gallagher-Jones, M. Dean, S. Wohlschlegel, J.A., Hill, K.L. (2021). APEX2 proximity proteomics resolves flagellum subdomains and identifies flagellum tip-specific proteins in Trypanosoma brucei. mSphere (In Press).
2.Walsh, B. & Hill, K. L. (2021). Right place, right time: Environmental sensing and signal transduction directs cellular differentiation and motility in Trypanosoma brucei. Mol Microbiol. doi:10.1111/mmi.14682.
3. DeMarco, S., Saada, E., Lopez, M., Hill, K.L.(2020). Identification of Positive Chemotaxis in the Protozoan Pathogen Trypanosoma brucei. mSphere5(4), e00685-20. https://dx.doi.org/10.1128/msphere.00685-20 . PMCID: PMC7426175.
4. Shaw, S.*, DeMarco, S.*, Rehmann, R, Wenzler, T, Florini, F., Roditi, I† and Hill, K.L.†(2019) Flagellar cAMP signaling controls trypanosome progression through host tissues. Nature Communications.10(1):803. doi: 10.1038/s41467-019-08696-y. *equal contributors; † co-corresponding authors
5. Imhof, S., Zhang, J., Wang, H., Bui, K., Nguyen, H., Atanosov, I., Hui, W., Yang, S., Zhou, Z., Hill, K.L.(2019). Cryo electron tomography with Volta phase plate reveals novel structural foundations of the 96-nm axonemal repeat in the pathogen Trypanosoma brucei. eLife 8, e52058. https://dx.doi.org/10.7554/elife.52058
6. Imhof, S. and Hill, K.L.Dynein-based motility of pathogenic protozoa, in: S.M. King (Ed.), Dyneins: Structure, Biology And Disease: Dynein Mechanics, Dysfunction, and Disease.vol. 2, Academic Press, Elsevier, (2018), pp. 418–435.
7. Shimogawa, M.M., Ray, S.S., Kisalu, N., Zhang, Y., Geng, Q., Ozcan, A., and Hill, K.L.(2018) Parasite motility is critical for virulence of African trypanosomes. Scientific Reports, 8, 9122. PMCID: PMC6002391
8. Zhang, Y., Ceylan Koydemir, H., Shimogawa, M.M., Yalcin, S., Guziak, A., Liu, T., Oguz, I., Huang, Y., Bai, B., Luo, Y., Luo, Y., Wei, Z., Wang, H., Bianco, V., Zhang, B., Nadkarni, R., Hill, K.L.and Ozcan, A. (2018) Motility-based label-free detection of parasites in bodily fluids using holographic speckle analysis and deep learning. Light: Science & Applications 7(1), 108. https://dx.doi.org/10.1038/s41377-018-0110-1. PMCID: PMC6290798
9. Langousis G, Shimogawa MM, Saada EA, Vashisht AA, Spreafico R, Nager AR, Barshop WD, Nachury MV, Wohlschlegel JA*, Hill KL* (2016). Loss of the BBSome perturbs endocytic trafficking and disrupts virulence of Trypanosoma brucei. Proc Natl Acad Sci U S A. 2016 Jan 19;113(3):632-7. doi: 10.1073/pnas.1518079113. (PMCID in progress) *co-corresponding authors
10. Shimogawa MM, Saada EA, Vashisht AA, Barshop WD, Wohlschlegel JA, Hill KL. (2015). Cell Surface Proteomics Provides Insight into Stage-Specific Remodeling of the Host-Parasite Interface in Trypanosoma brucei. Mol Cell Proteomics. 2015 Jul;14(7):1977-88. doi: 10.1074/mcp.M114.045146. (PMCID in progress). *co-corresponding authors.
11. Oberholzer M, Saada EA, Hill KL. (2015) Cyclic AMP regulates social behavior in African trypanosomes. MBio. Apr 28;6(3).doi: 10.1128/mBio.01954-14. (PMCID: PMC4436052)
12. Lopez MA, Saada EA, Hill KL. (2015) Insect stage-specific adenylate cyclases regulate social motility in African trypanosomes. Euk. Cell 14:104-12. doi: 10.1128/EC.00217-14. Epub 2014 Nov 21; PubMed PMID: 25416239. (PMCID:PMC4279026)
13. Saada EA, Kabututu ZP, Lopez M, Shimogawa MM, Langousis G, Oberholzer M, Riestra A, Jonsson Z, Wohlschlegel JA, Hill KL. (2014). Insect stage-specific receptor adenylate cyclases are localized to distinct subdomains of the Trypanosoma brucei flagellar membrane. Eukaryot Cell. 2014 May 30. EC.00019-14.
14. Freire ER, Malvezzi AM, Vashisht AA, Zuberek J, Saada EA, Langousis G, Nascimento JD, Moura D, Darzynkiewicz E, Hill K, de Melo Neto OP, Wohlschlegel JA, Sturm NR, Campbell DA. (2014) Trypanosoma brucei translation-initiation factor homolog EIF4E6 forms a tripartite cytosolic complex with EIF4G5 and a capping enzyme homolog. Eukaryot Cell. 2014 May 16. pii: EC.00071-14.
15. Freire ER, Vashisht AA, Malvezzi AM, Zuberek J, Langousis G, Saada EA, Nascimento JD, Stepinski J, Darzynkiewicz E, Hill K, De Melo Neto OP, Wohlschlegel JA, Sturm NR, Campbell DA. (2014) eIF4F-like complexes formed by cap-binding homolog TbEIF4E5 with TbEIF4G1 or TbEIF4G2 are implicated in post-transcriptional regulation in Trypanosoma brucei. RNA. 2014 Jun 24.
16. Kisalu, NK, Langousis, G; Bentolila, LA; Ralston, KS and Hill, KL. (2013). Mouse infection and pathogenesis by Trypanosoma brucei motility mutants. Cell Microbiol. 16(6):912-24. doi: 10.1111/cmi.12244.
17. Nguyen, H.T., Sandhu, J., Langousis, G. and Hill, K.L. (2013) CMF22 is a broadly conserved axonemal protein and is required for propulsive motility in Trypanosoma brucei. Euk Cell. 2013 Sep;12(9):1202-13.
18. Freund J.B.; Goetz, J.G.; Hill, K.L. and Vermot, J. (2012) Fluid flows and forces in development: functions, featrues and biophysical principles. Development 139(7):1229-45
19. Hughes, L. C., K. S. Ralston, Hill, K.L.*, Zhou, Z.H.* (2012). “Three-dimensional structure of the trypanosome flagellum suggests that the paraflagellar rod functions as a biomechanical spring.” PLoS ONE 7(1): e25700. doi:10.1371/journal.pone.0025700. *Co-corresponding authors.
20. Oberholzer M, Langousis G, Nguyen HT, Saada EA, Shimogawa MM, Jonsson ZO, Nguyen SM, Wohlschlegel JA*, Hill KL*. (2011) Independent analysis of the flagellum surface and matrix proteomes provides insight into flagellum signaling in mammalian-infectious Trypanosoma brucei. Mol Cell Proteomics. Oct;10(10):M111.010538. *co-corresponding authors
21. Ralston, Kisalu and Hill, KL. (2011) Structure-function analysis of dynein light chain 1 identifies viable motility mutants in bloodstream-form Trypanosoma brucei. Euk Cell (In Press – Journal Cover)
22. Merveille et al. (2011). CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat Genet. 43:72-78.
23. Kabututu, Z.P., M. Thayer, J.H. Melehani, and K.L. Hill (2010). CMF70 is a subunit of the dynein regulatory complex. J. Cell Sci. 123:3587-95.
24. Oberholzer, M.*, Lopez, M.A.*, McLelland, B.T. and Hill, K.L. (2010) Social motility in African trypanosomes. PLoS Pathogens, Jan 29;6(1):e1000739 (PMID: 20126443). *Co-1st authors.
-Selected as one of top ten news and noteworthy presentations at the 50th annual meeting of the American Society of Cell Biology (out of approx 4000 presentations).
-Covered by several news outlets, including radio interview for Voice of America (the official international broadcast service of the US).
-Selected as one of the top 42 most influential articles in the first decade of PLoS Pathogens.
25. Rodríguez, J. A., M. A. Lopez, M. Thayer, Y. Zhao, M. Oberholzer, D. Chang, N. K. Kisalu, M. L. Penichet, G. Helguera, R. Bruinsma, K. L. Hill and J. Miao (2009). “Propulsion of African Trypanosomes is Driven by Bihelical Waves with Alternating Chirality Separated by Kinks.” Proc Natl Acad Sci USA 106, 19322-19327. (PMID: 19880745. DOI: 10.1073/PNAS.0907001106).
26. Colantonio, JR, Vermot, J., Wu, D. Langenbacher, A., Fraser, S. Chen, JN and Hill, K.L. (2009). The Dynein Regulatory Complex is Required for Ciliary Motility and Otolith Biogenesis in the Inner Ear. Nature 457, 205-9 (2009).
